Past Research Areas:
CellProfiler and other bioimage analysis software
We launched and led the open-source CellProfiler project for 18 years. It is beloved by tens of thousands of biologists around the world. Since 2021, the Cimini lab is leading development (CellProfiler site).
CellProfiler Analyst: Machine learning for high-content screens
We launched and led the open-source CellProfiler Analyst project for 16 years. It helps biologists to train supervised machine learning algorithms to identify complex phenotype in high-throughput microscopy experiments. Since 2021, the Cimini lab is leading development (CellProfiler Analyst site) and working on Piximi to enable this kind of analysis using deep learning in a web browser.
Quantifying dynamic phenotypes
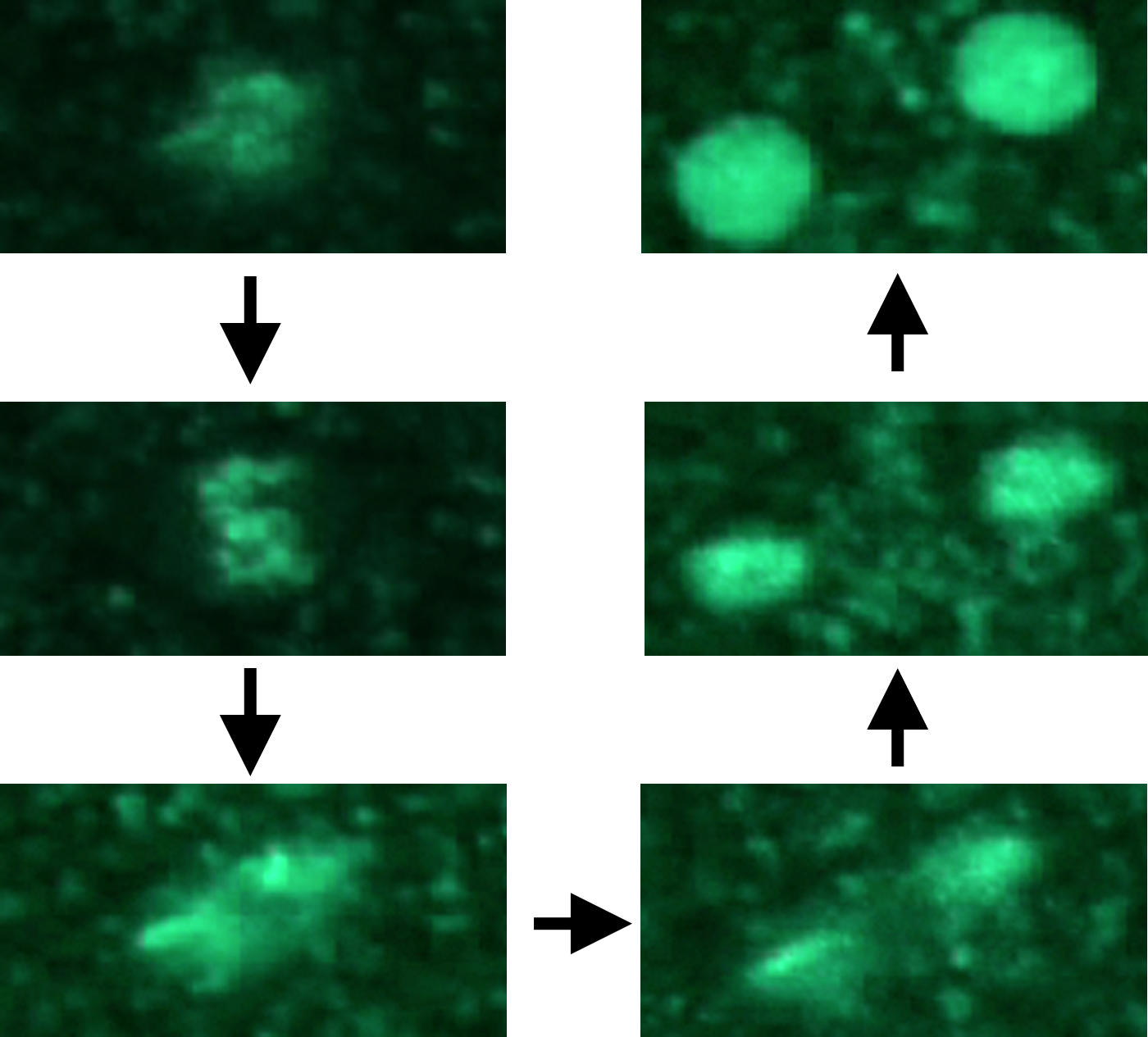 Many biological questions can only be investigated by collecting time-lapse movies. We have analyzed these images to identify, for example, novel cell cycle landmarks and motor protein regulators. We have also integrated this data with flow cytometry data to quantify unusual cell cycle outcomes.
Many biological questions can only be investigated by collecting time-lapse movies. We have analyzed these images to identify, for example, novel cell cycle landmarks and motor protein regulators. We have also integrated this data with flow cytometry data to quantify unusual cell cycle outcomes.
Imaging flow cytometry
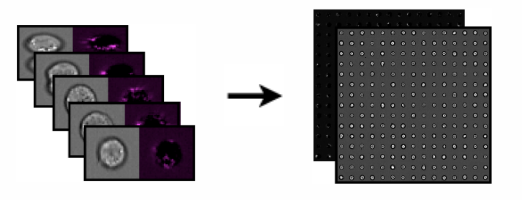 Imaging flow cytometry combines the high-throughput nature of flow cytometry with the high-resolution nature of fluorescence microscopy. For each experimental sample, it yields hundreds of thousands of images of individual cells. We developed methods to mine these large datasets [NSF project page] and demonstrated several novel applications where label-free imaging could replace customized fluorescence biomarkers.
Imaging flow cytometry combines the high-throughput nature of flow cytometry with the high-resolution nature of fluorescence microscopy. For each experimental sample, it yields hundreds of thousands of images of individual cells. We developed methods to mine these large datasets [NSF project page] and demonstrated several novel applications where label-free imaging could replace customized fluorescence biomarkers.
Co-culture systems
 In co-cultured cell systems, two or more cell types are grown together in order to maintain more native physiological functions, enabling experiments that test genetic and chemical perturbations in a more realistic environment. We developed image analysis approaches to extract information from fluorescence microscopy images of these cell systems, enabling experiments in liver regeneration and hepatotoxicity [NSF CAREER project page], and demonstrated several novel applications where label-free imaging could replace customized fluorescence biomarkers.
In co-cultured cell systems, two or more cell types are grown together in order to maintain more native physiological functions, enabling experiments that test genetic and chemical perturbations in a more realistic environment. We developed image analysis approaches to extract information from fluorescence microscopy images of these cell systems, enabling experiments in liver regeneration and hepatotoxicity [NSF CAREER project page], and demonstrated several novel applications where label-free imaging could replace customized fluorescence biomarkers.
Quantifying C. elegans
 The worm C. elegans can be robotically prepared and imaged and is an effective model to probe a variety of biological questions that require whole animals rather than isolated cells. We developed sorely needed C. elegans analysis algorithms and validated them in specific large-scale experiments to identify regulators of fat metabolism and pathogen infection.
The worm C. elegans can be robotically prepared and imaged and is an effective model to probe a variety of biological questions that require whole animals rather than isolated cells. We developed sorely needed C. elegans analysis algorithms and validated them in specific large-scale experiments to identify regulators of fat metabolism and pathogen infection.
 High-throughput imaging experiments generate extremely large, multidimensional data sets with quantifiable phenotypic information for every individual cell. Using machine learning, including deep learning, we mine this rich, latent information to identify patterns resulting from chemical or genetic perturbations to probe the causes and cures for various diseases. For example:
High-throughput imaging experiments generate extremely large, multidimensional data sets with quantifiable phenotypic information for every individual cell. Using machine learning, including deep learning, we mine this rich, latent information to identify patterns resulting from chemical or genetic perturbations to probe the causes and cures for various diseases. For example: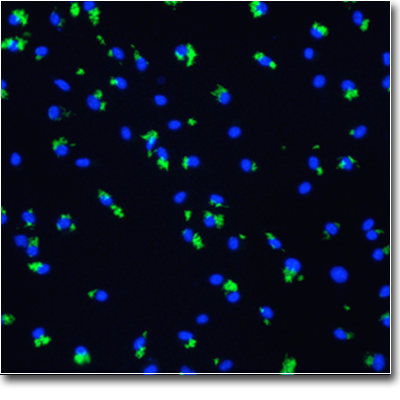 Our research has yielded discoveries in several translational projects, some of which have already had a direct impact on the treatment of disease. For example, we invented and maintained the CellProfiler project through 2021; it has been cited in over 20,000 papers. Discoveries made using CellProfiler have even led to clinical trials in humans and directly improved patient outcomes [
Our research has yielded discoveries in several translational projects, some of which have already had a direct impact on the treatment of disease. For example, we invented and maintained the CellProfiler project through 2021; it has been cited in over 20,000 papers. Discoveries made using CellProfiler have even led to clinical trials in humans and directly improved patient outcomes [

 Many biological questions can only be investigated by collecting time-lapse movies. We have analyzed these images to identify, for example, novel cell cycle landmarks and motor protein regulators. We have also integrated this data with flow cytometry data to quantify unusual cell cycle outcomes.
Many biological questions can only be investigated by collecting time-lapse movies. We have analyzed these images to identify, for example, novel cell cycle landmarks and motor protein regulators. We have also integrated this data with flow cytometry data to quantify unusual cell cycle outcomes. Imaging flow cytometry combines the high-throughput nature of flow cytometry with the high-resolution nature of fluorescence microscopy. For each experimental sample, it yields hundreds of thousands of images of individual cells. We developed methods to mine these large datasets
Imaging flow cytometry combines the high-throughput nature of flow cytometry with the high-resolution nature of fluorescence microscopy. For each experimental sample, it yields hundreds of thousands of images of individual cells. We developed methods to mine these large datasets  In co-cultured cell systems, two or more cell types are grown together in order to maintain more native physiological functions, enabling experiments that test genetic and chemical perturbations in a more realistic environment. We developed image analysis approaches to extract information from fluorescence microscopy images of these cell systems, enabling experiments in liver regeneration and hepatotoxicity [
In co-cultured cell systems, two or more cell types are grown together in order to maintain more native physiological functions, enabling experiments that test genetic and chemical perturbations in a more realistic environment. We developed image analysis approaches to extract information from fluorescence microscopy images of these cell systems, enabling experiments in liver regeneration and hepatotoxicity [ The worm C. elegans can be robotically prepared and imaged and is an effective model to probe a variety of biological questions that require whole animals rather than isolated cells. We developed sorely needed C. elegans analysis algorithms and validated them in specific large-scale experiments to identify regulators of fat metabolism and pathogen infection.
The worm C. elegans can be robotically prepared and imaged and is an effective model to probe a variety of biological questions that require whole animals rather than isolated cells. We developed sorely needed C. elegans analysis algorithms and validated them in specific large-scale experiments to identify regulators of fat metabolism and pathogen infection.