Guest Author
This post was written by a guest author, Kun-Hsing Yu, who can be reached at Kun-Hsing_Yu@hms.harvard.edu.
Lung cancer causes more than 1.4 million deaths per year. To diagnose lung cancer, pathologists prepare microscopic slides from surgical or biopsy samples, stain them with appropriate chemicals, and observe the visual patterns of cell morphology under the microscope. This manual (and often laborious) approach is the gold standard for lung cancer diagnosis and distinguished lung cancer subtypes.
Non-small cell lung cancer accounts for 85% of lung cancer. As the name suggests, the tumor is diagnosed by the microscopic morphology patterns of the tumor cells. Adenocarcinoma and squamous cell carcinoma are the two major subtypes of non-small cell lung cancer, and they are also defined by their visible cellular patterns under the microscope. Since many adenocarcinoma patients can benefit from targeted therapy, the distinction between these two subtypes is particularly important. However, studies have shown some level of inter-rater disagreements in classifying lung cancer subtypes using the conventional qualitative approach.
In addition, lung cancer patients have a wide range of survival outcomes, and it is difficult to predict patients’ prognoses with known clinical and pathological patterns. For example, more than 50% of the stage I lung adenocarcinoma patients passed away within 5 years despite standard treatment, but approximately 15% survived for more than 10 years. An accurate prediction of patient survival will not only guide clinicians to optimize treatment choices but also assist patients in advance care planning.
Survival outcomes of (1A) stage I lung adenocarcinoma and (1B) stage I lung squamous cell carcinoma patients. Tumor grade cannot reliably predict patient survival.
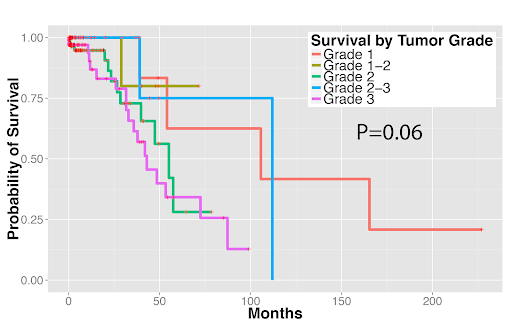
Figure 1A
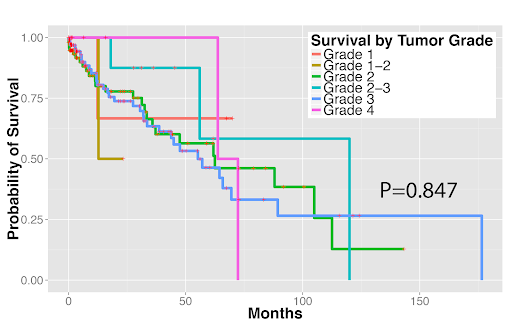
Figure 1B
Machine Learning for Pathology Diagnosis and Prognosis
In a published paper in Nature Communications, we used the CellProfiler package to segment the hematoxylin and eosin (H&E) stained pathology slides of non-small cell lung cancer patients, extract 9,879 objective features from the images, and correlate image features with pathology diagnosis and patients’ prognosis. Our results suggested the potential utility of quantitative pathology using CellProfiler. Below we describe our approach in detail.
We first used tools in the Open Microscopy Environment (OME) to convert the whole-slide pathology image files into manageable tiles, and selected the dense tiles that were likely to contain pathologic changes. We then used CellProfiler to segment the image and extract basic image features. Specifically, we used the ‘UnmixColors’ module to separate hematoxylin and eosin stains in the images, identified the cell nuclei by the ‘IdentifyPrimaryObjects’ module as well as the cell bodies by the ‘’IdentifySecondaryObjects’ module, and defined cytoplasm as the region within cell body but outside of nuclei. We extracted 790 features (with ‘Measure Correlation’, Measure Granularity’, ‘Measure Image Area Occupied’, ‘Measure Image Intensity’, ‘Measure Image Quality’, ‘Measure Object Intensity’, ‘Measure Object Neighbours’, ‘Measure Object Radial Distribution’, ‘Measure Object Size Shape’ and ‘Measure Texture’ modules) of the cell nuclei, cytoplasm, and the relations among them. We then summarized the distribution of cell-level quantitative features into 9,879 tile-level summary features.
Next, we employed supervised machine learning methods to distinguish tumor parts from adjacent dense benign tissue as well as to classify the two most common types of non-small cell lung cancer: adenocarcinoma and squamous cell carcinoma. Many machine learning classifiers achieved AUCs (areas under the receiver operator characteristics curves) of 0.73-0.88 in these classification tasks. And, importantly, the results were validated in an independent test set from the Stanford Department of Pathology.
Quantitative features distinguished tumor from adjacent benign tissues as well as tumor types. (2A) Receiver operator characteristics (ROC) curves for classifying lung adenocarcinoma from the dense adjacent benign tissue. (2B) ROC curves for classifying lung squamous cell carcinoma from the dense adjacent benign tissue. (2C) ROC curves for distinguishing lung adenocarcinoma from squamous cell carcinoma.
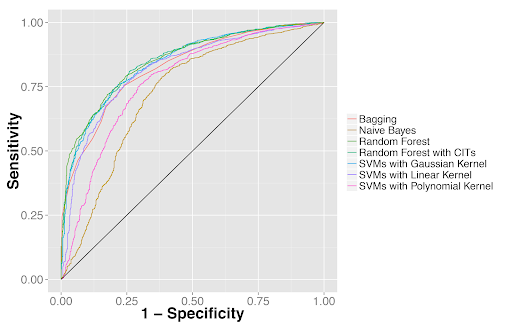
Figure 2A
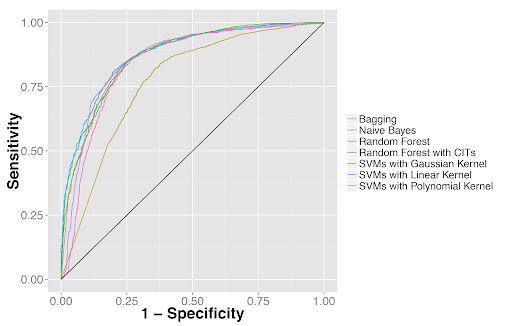
Figure 2B
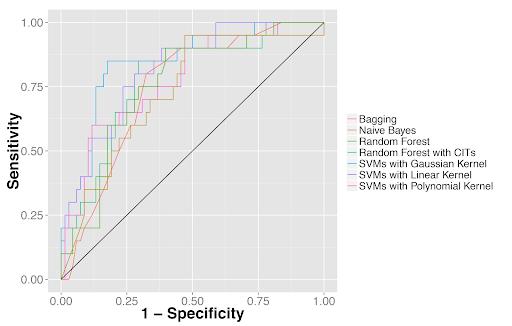
Figure 2C
Furthermore, we used the image features to predict patients’ survival outcomes in both tumor types. For stage I adenocarcinoma, we identified two survival groups with significantly different survival outcomes (P=0.0023). We also distinguished squamous cell carcinoma patients with different survival outcomes (P=0.023). Our approach was validated in the independent Stanford cohort (P=0.028 and P=0.035 for adenocarcinoma and squamous cell carcinoma, respectively). These results indicated the potential clinical utility of quantitative pathology evaluation.
Quantitative features distinguished longer-term survivors from shorter-term survivors with (3A) stage I lung adenocarcinoma and (3B) lung squamous cell carcinoma in the independent validation set from Stanford Department of Pathology.
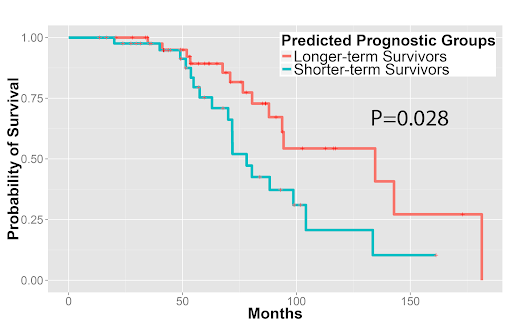
Figure 3A
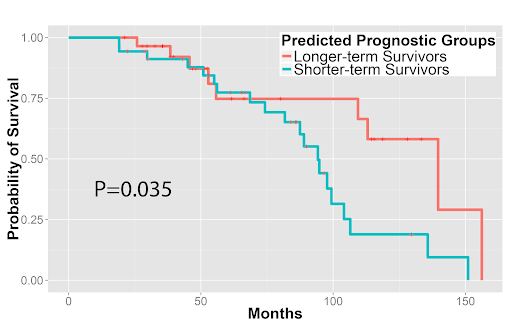
Figure 3B
CellProfiler for the Development of Quantitative Pathology Analysis
In our work, we found CellProfiler to be useful for processing H&E stained pathology images. It provides many helpful modules for unmixing the stains, identifying the cells, and extracting basic features. Stacking modules together forms useful pipelines for image processing and analysis. In addition, users can export the developed pipeline and run it parallelly for a large number of images. With the increasing availability of whole-slide pathology images and the growing computation power of parallel computing, quantitative pathology analysis can complement human evaluation and contribute to precision oncology.
The Paper Discussed in this Blog Post:
Yu KH, Zhang C, Berry GJ, Altman RB, Ré C, Rubin DL, Snyder M. Predicting non-small cell lung cancer prognosis by fully automated microscopic pathology image features. Nat Commun. 2016 Aug 16;7:12474. doi: 10.1038/ncomms12474.
Please find the full CellProfiler pipeline available for download here.
