Minh Doan
Thanks to the rapid advancement in image processing, we now have so many techniques to characterize cellular and subcellular objects (hooray CellProfiler!) Measuring cultured cells in monolayers is (usually) easy…but what about examining how cells interact with each other and their surroundings? Such experiments are often conducted using highly confluent cell cultures, tissue sections, or densely cell-packed organoids. At this level, clusters of cells gather, tightly bind and overlap to form cell niches, and often in a single area multiple clusters of various differentiated cell types can be found with different morphologies and functions. Recognizing and profiling individual cells can be very challenging under these circumstances.
Let’s look at some common challenges when analyzing tissue sections:
- Tissue sections are often stained with dyes (Hematoxylin and eosin, methylene blue, PAS etc.), that are not fluorescence-based. These absorbance-based dyes do not linearly nor quantitatively respond to the amount of material present. They are by nature invasive dyes that penetrate and stain homogeneously both cellular and extracellular materials, and some may contain heavy metals that are not suitable for fluorescence microscopy. In fact, some of these stains may enhance the appearance of extracellular matrix on purpose. Hence, cell junctions, cell adhesions, tissue fibers (e.g. collagens, elastic fibers), and other “background” are emphasized, not minimized, which does not help typical segmentation algorithms that work well on fluorescent stained images.
- A single image field may contain a complex mixture of very different and unique cell types, with different sizes and shapes, textures, and contents. Typical classical segmentation algorithms are often not “adaptive” enough to such dramatic changes, making it challenging to reach a high precision/accuracy in segmenting all different cell types in one image.
- Prior to staining, dissected tissue is often subjected to a harsh process: fixation, paraffin embedding, sectioning etc. These mechanical processes may cause certain materials to be lost. For instance, paraffin dissolves lipid, consequently lipid-containing regions (lipid droplets) of adipocytes in histology slides turn empty after being processed. At the same time, a tissue section may have some actual empty spaces in between, within and outside the cells (example below). So an automated segmentation looking for lipid droplets might be confused by these true empty spaces.
- Because overall “geography” is important, tissue sections are often imaged at relatively low resolution and/or with long working distance objectives to capture a wider view, in order to capture more cells per field. The details of the boundaries between cells are sometimes not well defined, and difficult to see even by eye.
The number of challenges may seem overwhelming, but don’t despair! In this post, we would like to discuss some solutions, or at least some ideas to facilitate the processing of tissue images.
- First, let’s start with sample preparation tips. Whenever possible, try to avoid the challenges described above – that is, use fluorescence dyes and image at sufficiently high resolution. Even better, include a DNA stain and if you can, include at least one good cell membrane stain in the preparation protocol.
- In the meantime, the following modules are those in our current version of CellProfiler that we have found to be particularly useful for tissue projects:
- UnmixColors (under parent category “Image Processing”). This module can be ‘trained’ to detect a particular color mixture that labels your cellular component of interest, or you can use pre-trained mixtures. Example
- ColorToGray (under parent category “Image Processing”), using its Conversion method “Split” to convert Red, Green, and Blue channels of the original images to gray; then you can use the ImageMath module to visualize and decide which combinations of channels would do best for your purposes. Example
- You may get the better results by combining the powers of different image processing software. Here we suggest:
- Orbit – open-source software that allows selecting regions of interest (perhaps manually) in huge format images so that smaller tiles therein can be processed in CellProfiler and returned to Orbit for viewing.
- ilastik – allows pixel classification. It takes into account not just the intensity levels of multiple channels but also the surrounding texture of each pixel to classify what cellular component each pixel belongs to.
Let us consider this histology image of brown adipose tissue:
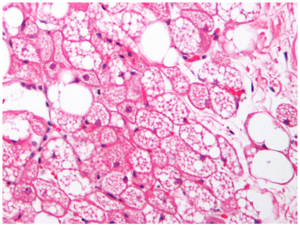
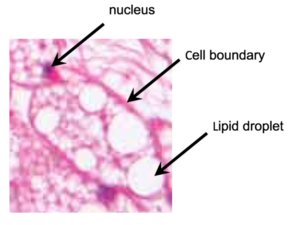
The nuclei are small and eccentric purple-blue specks, compared to the large red and white contents of the cells. The delineation between adjacent cells is a thick red line, but this can often be confused (even by experts) with the thinner red lines that delineate individual lipid droplets within each cell. The nuclei themselves are not particularly helpful for identifying individual cell regions; thus, the typical two-step strategy (identify nuclei first using IdentifyPrimaryObjects, and then use them as seeds to identify cell edges using IdentifySecondaryObjects) does not work well.
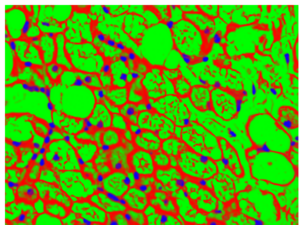
In this case, we can use ilastik as Kyle has demonstrated to generate a probability map of cell components, i.e. nuclei in blue, cell boundaries in dark red, and lipid droplets/ empty space in green:
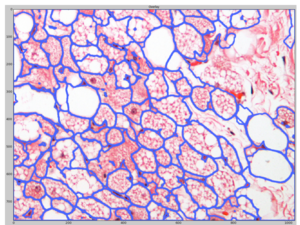
We then import the resulting probability map from ilastik into CellProfiler and use it to identify cells (marked by blue outlines):
In another example, thanks to a dedicated fluorescent stain for cell membranes, the recognition of individual cells can be accomplished with high accuracy:
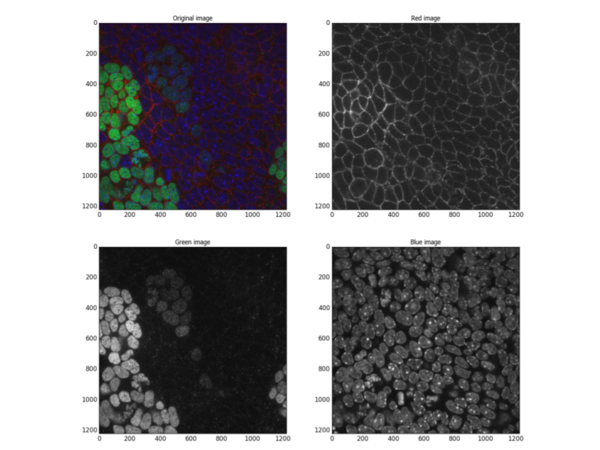
ilastik was then used to create a probability map of the membrane channel:
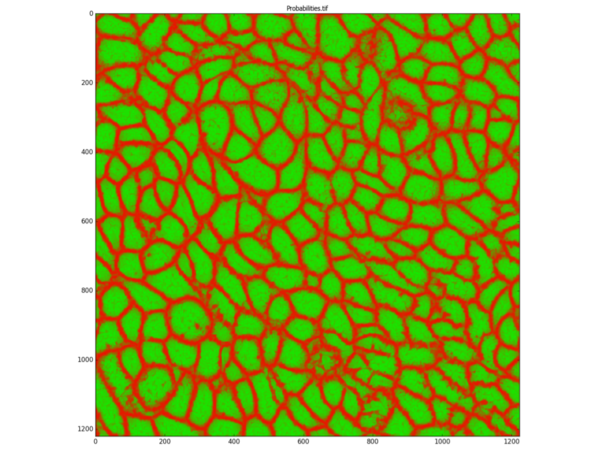
Fed by this probability map, CellProfiler can then successfully identify cells, skipping the need to even use the nucleus stain:
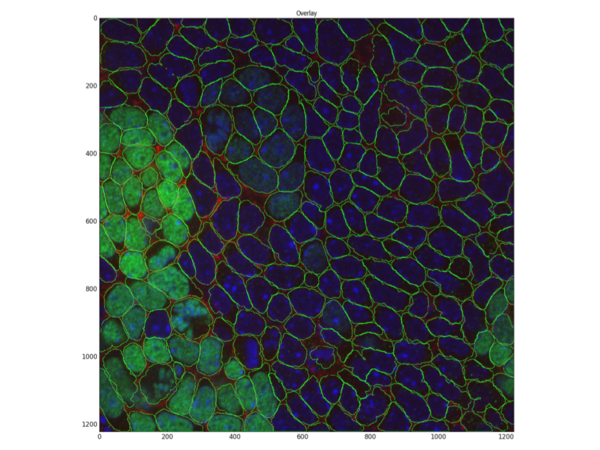
You can see how much a membrane stain can help in proper delineation of such challenging images of crowded areas.
Analyzing physiologically intact samples has a large and growing impact in biomedical research. Analyzing this type of image data is an area of active bioimaging research – we would love to hear your experiences and ideas for how to handle tissue samples.
