Nasim Jamali
As creators of open source tools, we spend a lot of time thinking about user needs - what kinds of images people are working with these days, what tools are working well, where the pain points are, and how easy it is to learn how to do any of this in the first place. While things like the image.sc forum give us a lot of insight into these kinds of questions, we also want to give people a chance to tell us this directly. We therefore teamed up with the Eliceiri group at University of Wisconsin in 2020 to put out a survey through our Center for OpenBioimage analysis (COBA), the results of which we recently published at Biological Imaging. These results will help not only the CellProfiler and ImageJ teams but the entire community, developers, trainers, and users as well as COBA like organizations to better prioritize their future tool development and educational programming.
In this post we are summarizing some of our favorite findings.
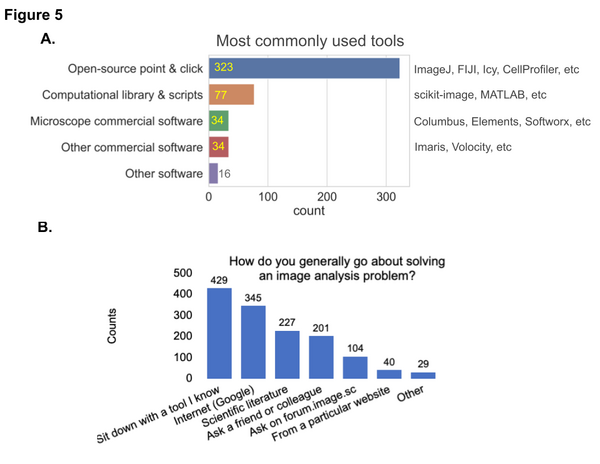
Commonly used approaches to solve image analysis problems
In general, the survey participants prefer to sit down with a familiar tool, search the web, look up solutions in the scientific literature, ask a friend or colleague, and/or ask on the Scientific Community Image forum (forum.image.sc)
Learning new topics and Method of learning
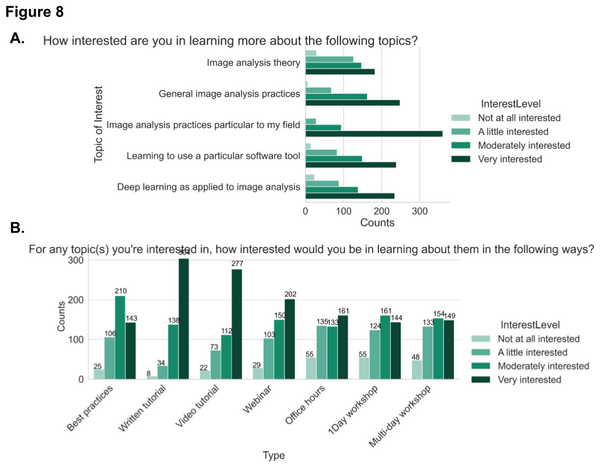
We wanted to find out from the community about the topics they are interested in learning and their preferred method of learning. The highest level of interest in learning about image analysis practices among the participants was specifically related to their own field of study/research. Written step-by-step and video tutorials are the top preferred methods of learning among participants.
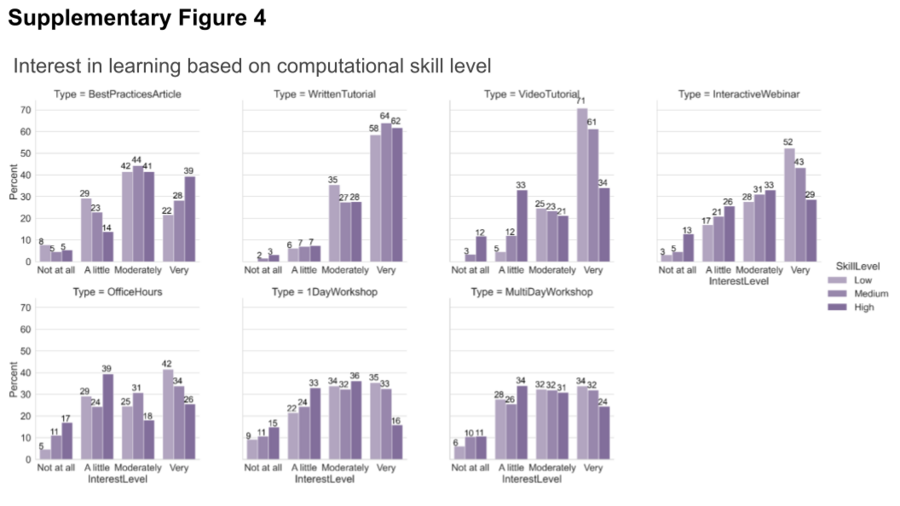
To further investigate the preference in various learning methods between different groups of survey participants, we broke down the results based on level of computational skills. We indicated that “the interest in written material remains high across all demographics but as the level of computational skills increases, the preference for best practices articles increases while the preference for video tutorials or more interactive offerings such as office hours and interactive webinars decreases”.
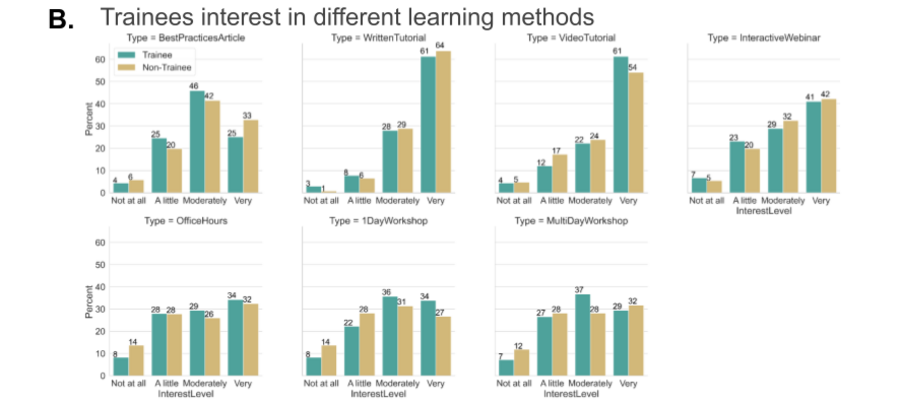
We also felt it was critical to assess the specific needs of trainees (grad students + postdocs) and their interest in different learning methods.
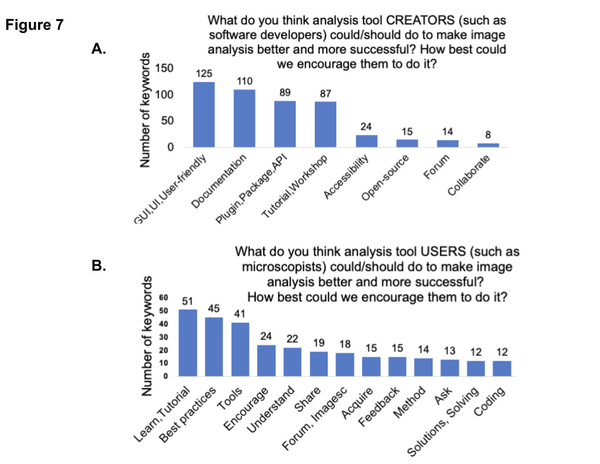
Community’s suggestions for the tool developers/creators and users
We then asked the community what they thought both users and tool developers could do to help one another: “the community’s requests for tool developers include general improvement of tool documentation and easy-to-follow tutorials. Respondents encourage tool users to follow the best practices guidelines for imaging and ask their image analysis questions on the Scientific Community Image forum (forum.image.sc)”.
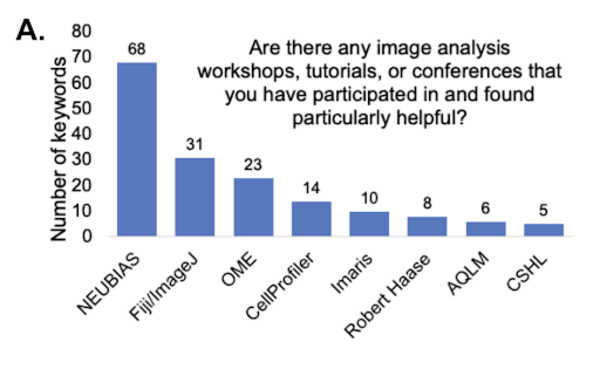
Helpful workshops and conferences
The community found the image analysis offerings from NEUBIAS (Network of European Bioimage Analysts), Fiji/ImageJ, OME (Open Microscopy Environment), CellProfiler, Robert Haase’s workshops on YouTube, Analytical and Quantitative Light Microscopy (AQLM) course at Marine Biology Laboratory, and the Quantitative Imaging course at Cold Spring Harbor Laboratory (CSHL), and Imaris workshops very helpful.
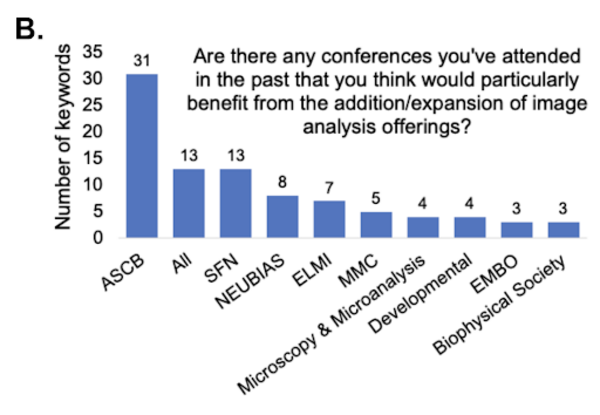
The community also suggested conferences such as the American Society for Cell Biology (ASCB), Society for Neuroscience (SFN), Network of European BioImage Analysts (NEUBIAS), European Light Microscopy Initiative (ELMI), the Microscience Microscopy Congress (MMC), Microscopy and Microanalysis, Developmental Biology, and European Molecular Biology Organization (EMBO) scientific conferences could benefit from the addition/expansion of image analysis offerings.
Thank you to all the co-authors (Beth Cimini, Ellen TA Dobson, Kevin W. Eliceiri, Anne E. Carpenter) and contributors. All the survey questions, responses to each individual question (sorted independently for each question to ensure deidentification), and all code used in creating the results are available here. If there’s anything else you want to know or questions you’d like to see in our next survey, please don’t hesitate to reach out via contact forms on our website (https://openbioimageanalysis.org/), email (COBA@broadinstitute.org), or COBA Twitter account: @COBA_NIH).
