Beth Cimini
Defining the input to CellProfiler can be the hardest part of getting your pipeline set up and your analysis underway. Incoming images are configured in the first 4 modules of CellProfiler – Images, Metadata, NamesAndTypes, and Groups – which offer lots of flexibility. But it’s sometimes confusing what each one does, and it’s not always obvious which ones you need for your experiment.
If you have mistakes in any of these modules, you may run across the dreaded errors ‘The pipeline did not identify any image sets’ or ‘Sorry, your pipeline doesn’t produce any valid image sets as currently configured’. These errors simply mean that CellProfiler doesn’t know how to sort the images you’ve given it, and it can’t process them until it knows how they should be organized.
If you want to learn more, we recommend reading the help sections for each module, and everything under ‘Help->Creating A Project’ is a fantastic resource as well (we’ve included a table from it below that’s a great cheat sheet to understanding these modules). But here are some common triggers for this kind of error.
- Your image files have been moved to a different location on your computer, but CellProfiler is still looking for them in their original location.
- You forgot to hit ‘Update’ in the Metadata module when using the ‘Extract from image file headers’ option.
- A piece of extracted metadata is missing (e.g., you remembered to extract the Plate metadata from your ‘regular’ images but not from your illumination correction functions). Check the output of the Metadata module to ensure all of your images have the information that they need.
- You turned on ‘Groups’ but never gave it a piece of metadata to group by (in which case the ‘Metadata category’ is still set to ‘None’).
- One of the sets of rules you’ve used to identify a channel isn’t set correctly – for example:
- You’ve told it to set files containing ‘ch4’ as your DNA image but your experiment only has files ending in ‘ch1’, ‘ch2’, and ’ch3’.
- You’ve told it to use ‘Directory does contain ‘ch3’’ to define your DNA image, but you meant to set it to ‘File does contain ‘ch3’’.
- You’ve used ‘Metadata’ for your Image set matching rather than ‘Order’, but you haven’t set it up correctly.
These last two errors are particularly sneaky: if you hover over a dropdown menu with a scroll-wheel mouse, you can accidentally scroll down to a different option, messing up a configuration that was previously correct. We’re working to try to correct this problem in future versions of CellProfiler.
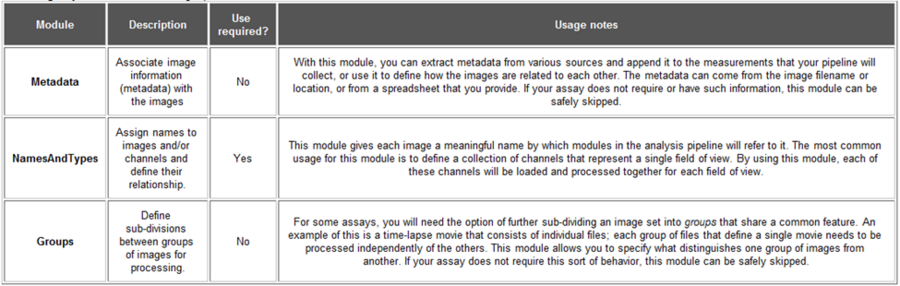
Table 1: A quick guide to the functions of Metadata, NamesAndTypes, and Groups (from Help->Creating A Project->Configuring Images for Analysis)
Did these tips help you? Are you having trouble configuring a CellProfiler pipeline? Did you have a tricky configuration issue that you finally solved? Tell us below!
