Kyle Karhohs
Now that CellProfiler can handle 3D images, how do you view the results? When processing 2D images, the IdentifyPrimaryObjects module will produce a figure that displays the original image next to the segmented image to make validation fast and convenient, a side-by-side comparison. However, the extra dimension in 3D complicates this approach, because the 3D image must be transformed in some manner in order to appear on a flat surface (e.g. the monitor or phone you’re using to read this blog post!). This blog post contains advice on how to look at 3D images and the 3D segmentation image produced by CellProfiler and introduces orthoviews in FIJI. If you would like to explore the example used in this post please check out the demo posted on the CellProfiler forum.
The output figure created by 3D capable modules in CellProfiler will show up to 9 slices of the volume image (see the figure below). Tiling the slices of an image in this manner is a convenient form factor for displaying a large amount of information in a single view; all at once we can see what is happening in the top, middle, and bottom of the volume. Segmentation can be validated by eye in much the same way as for 2D images: the tile pair that corresponds to the input image and the segmented image can be compared side-by-side.
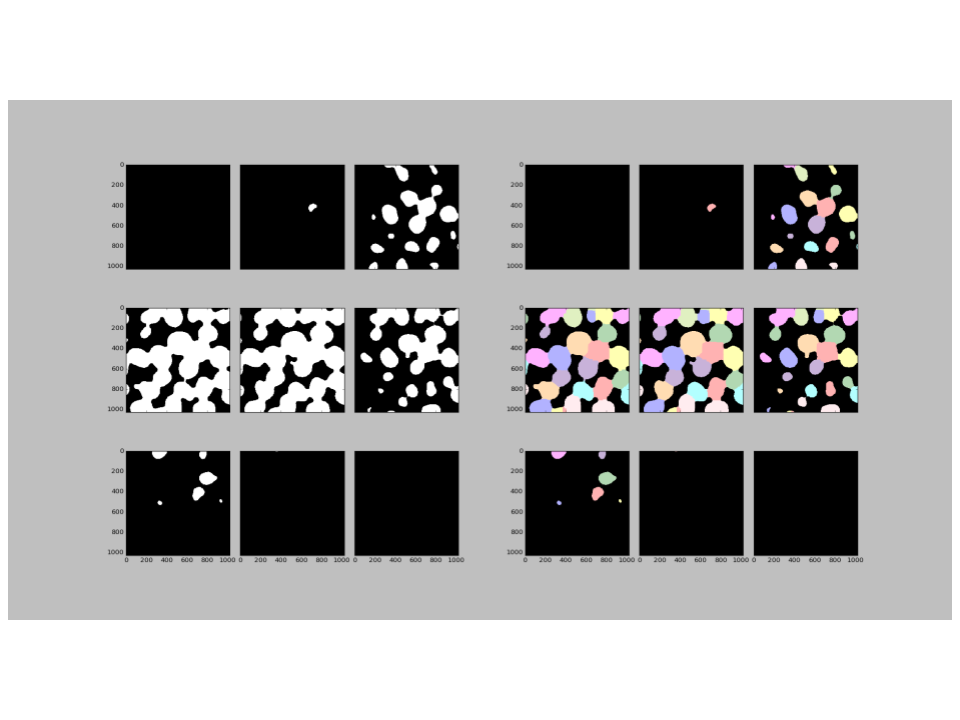
Tiled slices are effective at identifying gross errors in the segmentation, such as widespread over- or under-segmentation. However, the tiled slices of a volume start to become less effective as more tiles are displayed. Eventually, tiles can appear crowded and small, so if you’re interested in the segmentation of one object in particular or optimizing a threshold it can be difficult to see changes. We are also limited to a top-down or XY view of the 3D stack and might miss an important detail that would be clearly seen from a different perspective.
FIJI is an open-source tool based on ImageJ2 that creates interactive orthoviews of a 3D stack to see the finer details of segmentation. To activate the orthoview follow the menu tree: Image > Stacks > Orthogonal Views. To prepare your image for FIJI, use the OverlayOutlines module in CellProfiler to overlay object outlines onto the original image; remember to save the result using the SaveImages module. An overlay will reveal exactly where the segmentation and source image agree or disagree. The orthoviews in FIJI provide an interactive way to evaluate a CellProfiler pipeline (see the movie below). The motion of scrolling through a Z stack can be reveal changes that might be otherwise be overlooked in a static slice. FIJI is thus a great open-source option to help visualize the segmentation of 3D image data when you need something more powerful than CellProfiler’s tiled view.

*Note: The upcoming release of CellProfiler 3.0 will add functionality to segment volumes, i.e. stacks of images with 3 dimensions X, Y, and Z. This 3D functionality can be tested in advance of the official release by running CellProfiler from source. Please visit Github wiki for instructions on how to install from source.
