Microscopy in the cloud: CellProfiler and Cell Painting on Terra
Authors: Carmen Diaz Verdugo, Stephen Fleming, Nicole Deflaux and Amy Unruh
Carmen Diaz Verdugo is a Computational Scientist in the Precision Cardiology Lab, and a member of Patrick Ellinor’s group. Stephen Fleming is a Senior Machine Learning Scientist in the Methods group of the Data Sciences Platform at the Broad Institute, and he is also a member of the Precision Cardiology Lab. Nicole Deflaux and Amy Unruh are software engineers at Verily Life Sciences and members of the Terra team.
In this guest blog post, Carmen, Stephen, Nicole, and Amy introduce several cloud-optimized workflows for running CellProfiler and Cell Painting pipelines at scale on Terra, the secure biomedical research platform co-developed by Broad Institute of MIT and Harvard, Microsoft, and Verily. The workflows live in a publicly-accessible Broad GitHub repository and are hosted on Dockstore.
Image-based cell profiling
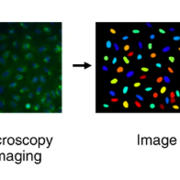
Image-based – or “morphological” – profiling combines high-content image-based assays, such as the Cell Painting assay, with computational methods to analyze the resulting data. The Cell Painting assay uses a collection of fluorescent dyes that are multiplexed in different channels to reveal the most relevant cellular compartments or organelles such as the nuclei, cytoskeleton or mitochondria. Cells are placed in multiwell plates, stained, perturbed either genetically or with compounds, and imaged with a high-throughput microscopy system. Next, using image analysis software (for which we use the open-source software CellProfiler), cells are identified, segmented, and thousands of morphological features are measured. The collection of features is known as a profile, and it describes the morphological properties of the cells treated with the specific perturbations.
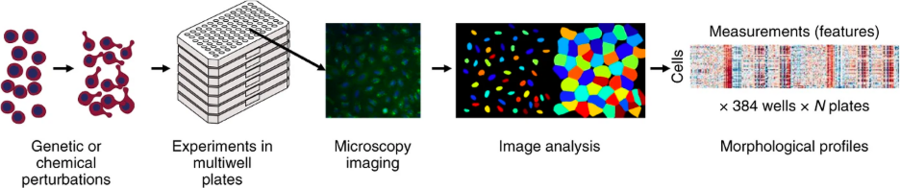
From: Bray, MA., Singh, S., Han, H. et al. Cell Painting, a high-content image-based assay for morphological profiling using multiplexed fluorescent dyes. Nat Protoc 11, 1757–1774 (2016) [https://doi.org/10.1038/nprot.2016.105]
Cell Painting is an emerging valuable tool, and several labs in academic settings and pharma companies use this assay, in large part because of its several applications in the drug discovery pipeline. The Broad Institute of MIT and Harvard, where the Cell Painting assay protocol was developed, launched a collaboration with industry and non-profit partners called Joint Undertaking in Morphological Profiling with Cell Painting (JUMP-CP). This consortium recently released a large collection of Cell Painting datasets covering 136,000 chemical and genetic perturbations open to the scientific community for data exploration. The CellProfiler and Cell Painting workflows in Terra that will be described in this blog are tools to facilitate the analysis and exploration of these large datasets using cloud services.
Image analysis with CellProfiler
CellProfiler is open-source software for measuring and analyzing images, designed with an intuitive user interface that allows scientists to analyze large amounts of microscopy images automatically. The CellProfiler project was started by Anne E. Carpenter and Thouis (Ray) Jones and it is currently based in the Cimini Lab at the Broad Institute.
CellProfiler can read and analyze most common microscopy image formats, and the software offers a wide range of modules for image preprocessing, object/cell segmentation, and measuring quantities such as area shape, intensity, and texture. The measurements are exported as CSV files or a SQLite database.
The need for scale
Currently, with the advances in automated high-content microscopy systems and robotic equipment in laboratories, lab scientists are able to generate large amounts of data that are challenging not just to analyze, but also to store. Research labs and pharma companies alike are running up against a potential bottleneck: running the software pipelines to extract CellProfiler features can take longer than collecting the actual data, not to mention all the downstream analyses that follow CellProfiler feature extraction. Running CellProfiler pipelines sequentially on a single computer is not scalable. Bespoke solutions using multiple compute nodes on local computer clusters can work, but require a great deal of skilled development and maintenance, and involve a large learning curve. That’s where parallel compute in the cloud can offer a truly scalable alternative for those with lots of data to process.
A cloud-based solution
Terra is a scalable platform that allows researchers to access data, run analyses, and collaborate using secure cloud services. We have created a series of WDL workflows that allows you to run CellProfiler, for either Cell Painting pipelines or any custom pipeline, and PyCytominer tools, which are a collection of functions to process high-dimensional readouts from high-content experiments. These pipelines can be run with very little setup at the click of a button. Compute capacity will automatically scale with the size of the dataset, performing the analysis in parallel, and Terra will email you when the job is complete. The WDL workflows include three pipelines – “CellProfiler”, “Cell Painting”, and “Cytomining”:
- CellProfiler pipeline is a basic workflow that runs any custom .cppipe CellProfiler pipeline on Terra. The pipeline uses the LoadData input module can be specified as usual for a headless CellProfiler run. Just pass the .cppipe file and a path to the relevant images in a Google Cloud Storage bucket, specify how to split up the workload, and the workflow will spin up one or more virtual machines (VMs) on Google Cloud, per your specifications, and run the CellProfiler pipeline there. Output can either be a tarball (called "output.tar.gz", and located wherever Terra chooses) or the output files can optionally be extracted and copied to a Google Cloud Storage bucket location of your choice (see the optional input output_directory_gsurl).
- Cell Painting pipeline contains four workflows to be run sequentially in order to run a full end-to-end Cell Painting analysis:
1. create_load_data: This workflow creates load_data.csv and load_data_with_illum.csv files that CellProfiler uses for loading each of the images for analysis. Users can skip this step if they opt to create those files either manually, exporting the image set list using the CellProfiler GUI, or using other available resources (e.g. pe2loaddata).
2. cpd_max_projection_pipeline: This workflow runs the CellProfiler maximum intensity projection pipeline in a distributed fashion by running it on multiple VMs simultaneously. This workflow is optional, and it is meant to be used when more than one plane of the fields of view have been acquired.
3. cp_illumination_pipeline: This workflow runs the CellProfiler illumination correction pipeline on a single VM (not distributed).
4. cpd_analysis_pipeline: This workflow runs the main CellProfiler pipeline in a distributed fashion, using multiple VMs in parallel, which typically performs cell segmentation and measures Cell Painting features.
- Cytomining pipeline runs the cytominer-database ingest step to create a SQLite database containing all the extracted features, and runs the aggregation step from pycytominer to create CSV files.
These workflows are all publicly available, and hosted in Dockstore. You can import and run the workflows in Terra or any other place you like to run WDL workflows.
Get started with CellProfiler on Terra
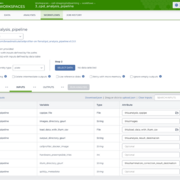
If you are excited to try these workflows, we have prepared a Featured Workspace in Terra, https://app.terra.bio/#workspaces/cell-imaging/cellpainting, that you can clone and use to run an example of a Cell Painting analysis step by step! It includes a Terra Data Table, demonstrating how to run these workflows on multiple plates simultaneously, if desired.
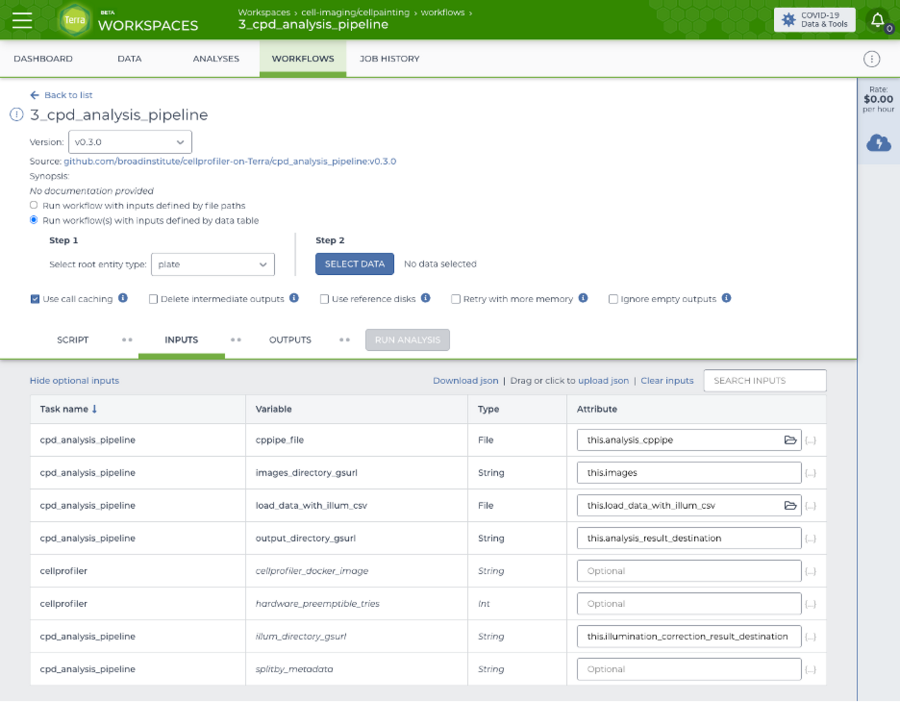
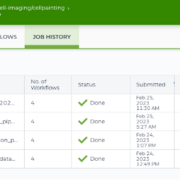
Screenshot of the Terra workflow page used to launch a CellProfiler analysis on several plates in parallel.
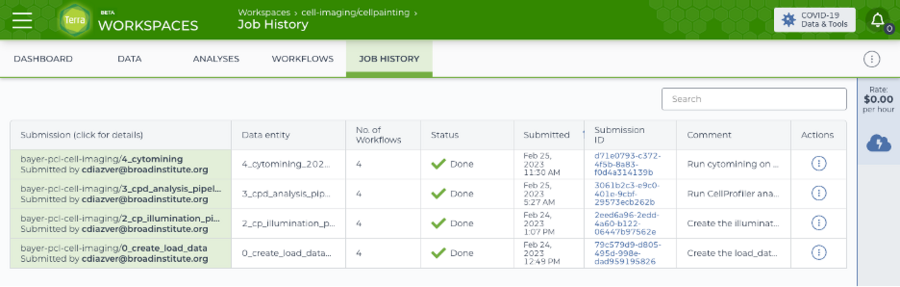
Screenshot of the completed runs of all four workflows to create the load data files, correct illumination, analyze via CellProfiler, and then aggregate the features using pycytominer.
Using Terra to run CellProfiler pipelines in the cloud offers researchers a scalable and cost-effective solution for processing and analyzing large volumes of high-content microscopy data. We hope that these publicly available workflows will be of interest to the research community, and that they will facilitate Cell Painting and other morphological profiling analyses at scale.
Resources