Melissa Gillis
Segmenting fibers using traditional cell segmentation software can be challenging. Because most biological cell segmentation tools were designed for cells, most programs have difficulty identifying and distinguishing longer and thinner objects such as fibers. Also, unlike cells, fibers tend to overlap with one another, making it difficult to distinguish individual fibers from one another. A versatile model that can accurately distinguish and segment fibers has yet to be developed. In this series I will explain how I created a model that successfully segmented a variety of fiber types.
Previously, Pearl Ryder had written a series of blog posts titled “Thinking like an Image Analyst”. In this series, Pearl described how she used CellProfiler to segment fibronectin fibers in images acquired by the Gaborski NanoBio Materials Laboratory at the Rochester Institute of Technology. Tom Gaborski’s Laboratory studies how porous substrates affect the extracellular matrix, which has important consequences for cell migration. Pearl originally worked with immunofluorescence microscopy images originally published by Chung et al. in ACS Biomaterials Science & Engineering. These images presented multiple challenges: 1) They were very dim and had low contrast making it difficult to distinguish fibers from background. 2) Many of the images contained bright circular debris irrelevant to the fiber analysis. Examples of these images are shown below.
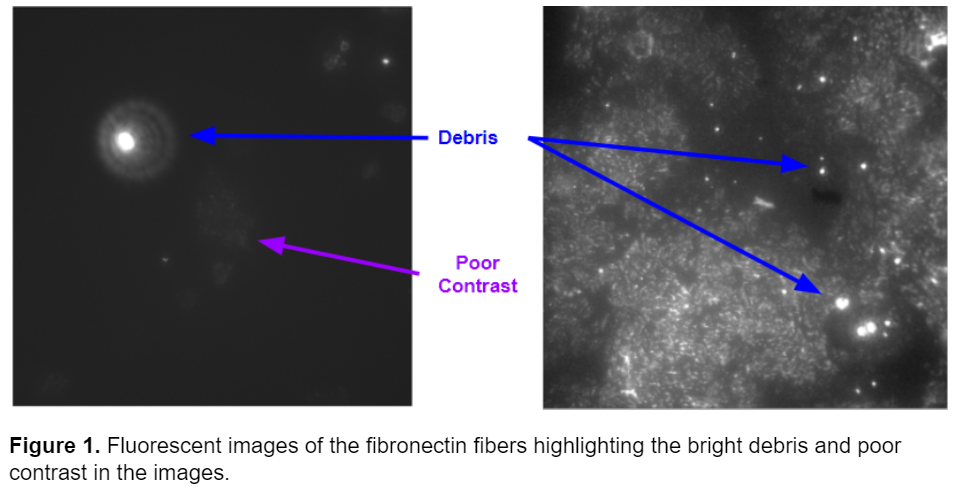
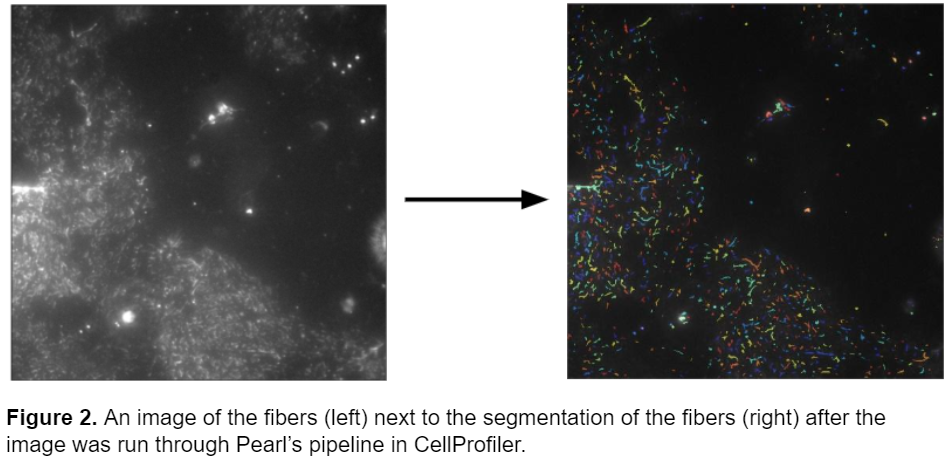
In her blog series, Pearl explains how she removed the debris in the images and corrected for the poor contrast. Her pipeline was very successful at accurately segmenting fibers. Below a raw image of the fibers is compared to the segmentation after the image is run though Pearl’s pipeline. The pipeline accurately segments the fibers and eliminates the bright debris in the image.
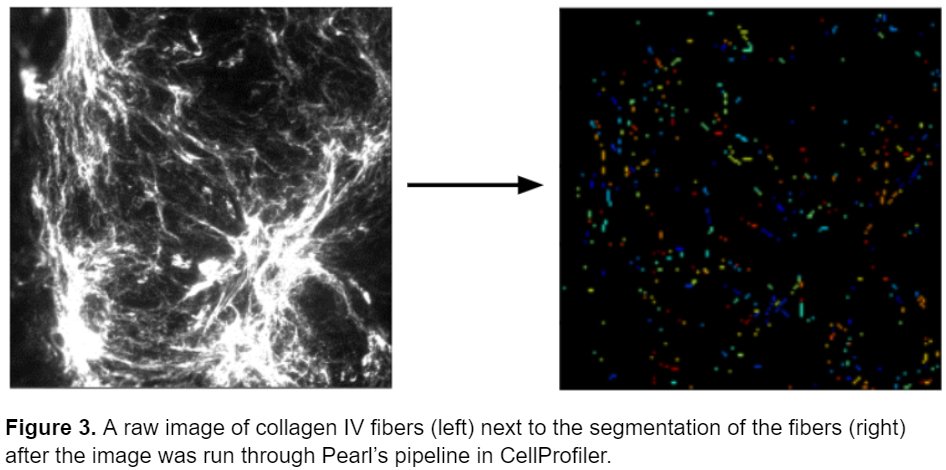
However, while the pipeline performed well on this specific type of fiber, it wasn’t as successful at identifying different types of fibers in different conditions. Below is an image of collagen IV fibers taken from the same lab with a slightly different experimental set up a few years later. Unfortunately, this pipeline fails to accurately represent the morphology of the fibers and represents them as small elliptical objects instead of mimicking their thin and elongated shape.
While Pearl’s pipeline was very successful at segmenting the original fibers, it was too specific to accurately segment any other type of fiber. Therefore, I set out to create a single model that could accurately segment a variety of fiber types eliminating the need for multiple models.
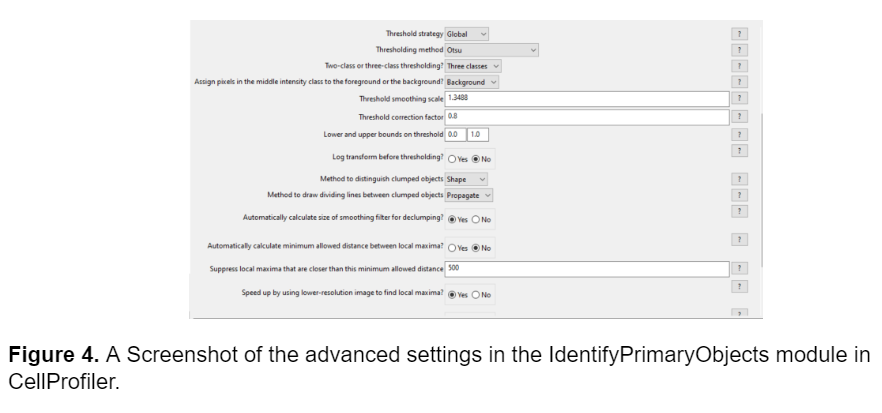
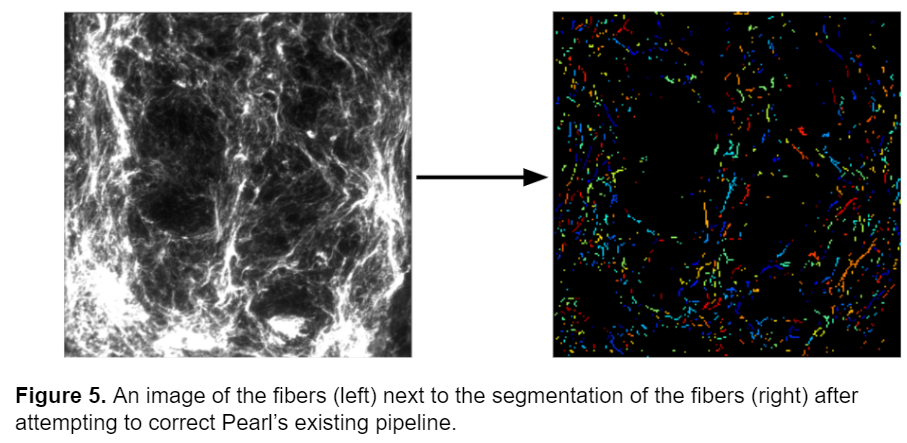
Initially, I tried to adjust Pearl’s existing pipeline to generate an accurate segmentation model. The key module in Pearl’s pipeline that segmented the fibers and differentiated them from the background was the IdentifyPrimaryObjects module, so I started by adjusting the advanced settings. I tried adjusting the minimum allowed distance between local maxima (which determines the necessary proximity between pixels to be counted as separate objects), adjusting the size of the smoothing filter (which blurs or sharpens the fibers in the image before segmenting), and changing the thresholding method, but these adjustments didn’t prove sufficient in accurately segmenting the fibers. The model was still unable to identify dimmer fibers and the segmentation wasn’t an accurate representation of the morphology of the fibers. A screenshot of the advanced settings in the IdentifyPrimaryObjects module as well as an example of the segmentation is shown below.
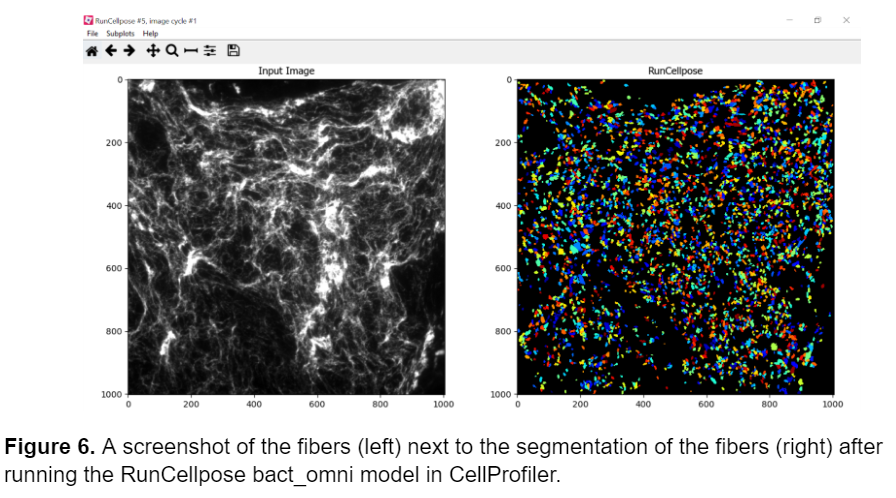
Next, I tried using the RunCellpose module in CellProfiler in hopes that it would serve as a better model. RunCellpose runs the Cellpose deep learning models in CellProfiler, which can perform better with challenging segmentations. I initially started with the bact_omni model that uses omnipose for segmentation because it performs well with segmenting long, thin bacteria that are similar in morphology to the fibers. Unfortunately, it didn’t work as I had hoped, and the fibers were still segmented as circular objects (image shown below). I also tried the other Cellpose models on the fibers including cyto, nuclei, and even attempted to create a custom model using the cellpose 2.0 app, but all proved insufficient.
After exhausting the options available in CellProfiler, I considered using a separate software to generate probability maps of the fibers and then import those maps in CellProfiler to segment and measure the fibers. I decided to use a machine learning model to detect the fibers within each image. I attempted using the lab’s deep learning web application Piximi to annotate the images and then export them as label matrixes. Initially, annotating each image by hand seemed reasonable, however the density and interwoven pattern of the fibers made it very challenging to determine where individual fibers started and ended, making individually annotating all the fibers in an image impossible. I knew that I needed to develop a machine learning model that could identify the fibers in these images and sought out to explore training this model in ilastik. In the next section of this series I will explain how I used ilastik to accurately segment the fibers.
Customizing a Model for Fiber Segmentation Series:
Part I: Investigating Possible Methods
Part 2: Creating an ilastik Model
Part 3: Constructing a CellProfiler Pipeline
Part 4: Analyzing the Results
