Anne Carpenter
 So you already know how to put together an image analysis pipeline to measure particular phenotypes of interest? Great!
So you already know how to put together an image analysis pipeline to measure particular phenotypes of interest? Great!
Have you ever considered looking for the unexpected? Say you are comparing two treatment conditions, such as a negative control vs. a hormone treatment. You may have in mind phenotypes to measure, so you use CellProfiler to accurately The
quantify them. But did you realize you could also measure everything you can from the images and let the data tell you what distinguishes your two conditions?
Here at the Broad Institute’s Imaging Platform, our informal motto is “Measure everything, ask questions later”. We took this approach in the original CellProfiler paper. We wanted to measure translocation, but on a whim measured many more parameters. We uncovered an unexpected change in nuclear morphology! In a subsequent paper, we outlined a CellProfiler-friendly protocol for taking this systematic approach for finding differences between two sets of samples (e.g., negative control vs. treatment).
We are taking this a step further in experiments involving patients with psychiatric disease, where the molecular pathways involved are murky and therapies are desperately needed. Taking groups of patients with and without a disorder, we are measuring everything we can about the cells and let the data tell us what distinguishes them.
Now, is this approach literally “unbiased”? Well, no: you’ve selected a specific cell type, set of stains, timepoint, imaging conditions, and so on, so you will only detect morphological changes visible under those conditions. And, you need to choose which cellular regions to identify (segment) in the images (unless you go for deep learning – more on that in future posts!) Further, you can add as many Measure modules as you like, but you won’t be adding an infinite number and even if you did, these would not fully capture every bit of information in images.
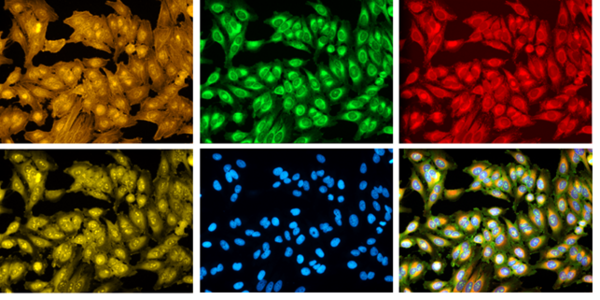
Nevertheless, you can make an experiment more unbiased, for example, by adding as many stains as possible. We developed the Cell Painting assay for this: it’s an easy protocol with six inexpensive stains that capture a wide variety of morphological information about organelle structures. Methods for sequential staining/bleaching and imaging mass spectrometry could provide even more information!
Are there any downsides to this approach? Well the more cellular features you measure, the more opportunity to find a morphological difference by chance alone and fool yourself. This is a common problem in machine learning: beware of Voodoo machine learning (warnings described in Logan, Shamir, Saeb).
The vast majority of “high-content screens”, although named for their high information content, only measure 1-2 features of cells (Singh paper). Taking an unbiased approach based on measuring hundreds of features… who knows what you will see!
We’d love to hear examples of when you went looking for the unexpected in images, and what you found.